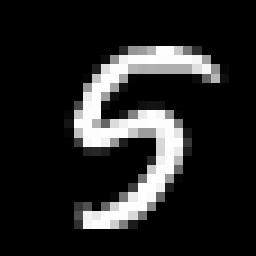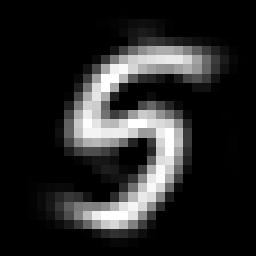So far, k-NN has turned out to be a reasonably accurate, if rather slow performing classifier on the MNIST dataset. At over 20 minutes to compute the results for the test data set on my iMac, and even longer when one takes into cross-validation for debugging on training data, it's clear that such a research approach isn't sustainable.
Example 1's from the training data set. Notice the wide variety of examples and how they are transformed: rotation, translation, fattening, etc.
How might this be remedied? One possibility is through data compression. By compressing the image data in much the same way as everyday image files are compressed, we can greatly reduce both the amount of computation and space required to perform k-NN.
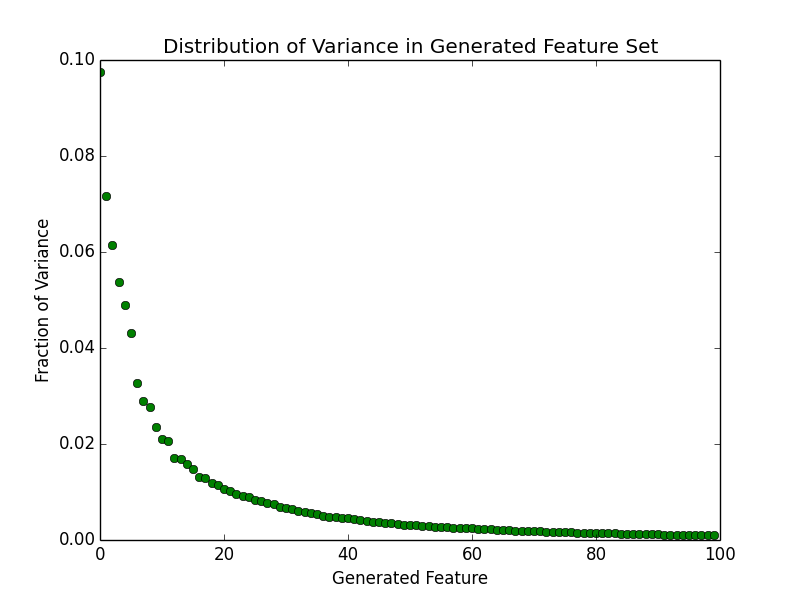
Previously, we saw that the distribution of variance across pixels is fairly wide. If there were some way to create a new feature set in which the variance is compressed into a small sub-set of the features, then we might be able achieve our goal. As it turns out, such techniques do indeed exist. The one we are interested in today is called Principal Component Analysis (PCA). This is a technique based on linear algebra. I'm not going to describe it in detail today, but the major idea is that it finds a new set of basis vectors in which most of the variance falls within or near a small number of dimensions. Shown below, is the distribution of variance for the training examples for the first 100 features.
Most of the variance seems to be compressed in the first 40 or so features. This is a big improvement. Instead of dealing with 784 dimensions we are probably looking at less than one tenth that number. It's important to keep in mind that these new features don't have any meaning in normal everyday terms. They certainly don't correspond to 'pixels' anymore. Also, the compression process is lossy for any number of new features less than our original number, so we are losing some information. As shown below, reconstructing an example digit from the new basis results in a digit that is "fuzzier".
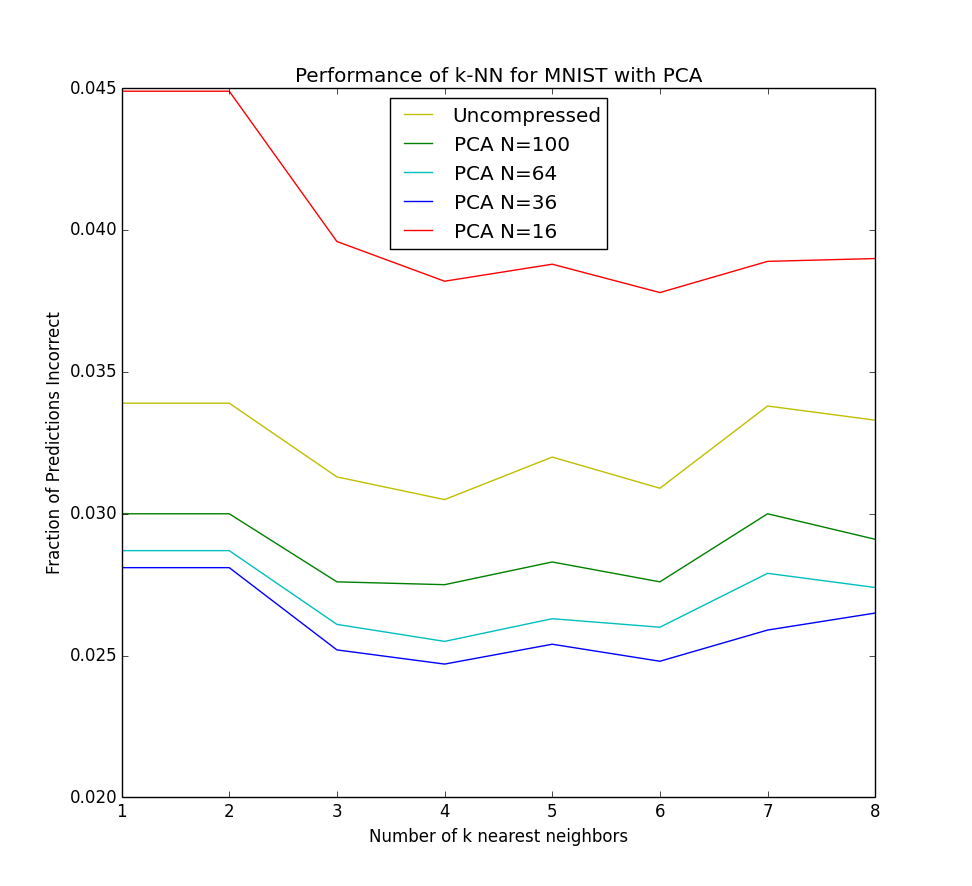
So how did we do on the training data? After trying a few different values for the number of PCA components, N, I settled on a value of 36 as being quite good. This value resulted in an execution time of around 37 seconds, a big improvement from before! This allowed me to rapidly test a number of configurations for both N and k. Going to significantly fewer dimensions however (16 for example), had the opposite effect and actually hurt accuracy. The results on the cross validated training set are shown below.
Surprisingly (or perhaps not), it turns out that not only did using PCA improve the speed of computation, it also resulted in greater accuracy! My submission from this round of tweaks came in at a respectable 97.5% accuracy. Hooray!
In upcoming posts, I plan to delve into the details of how PCA actually works and why it's possible to not only get a speed improvement but accuracy as well by effectively removing variables from the data.